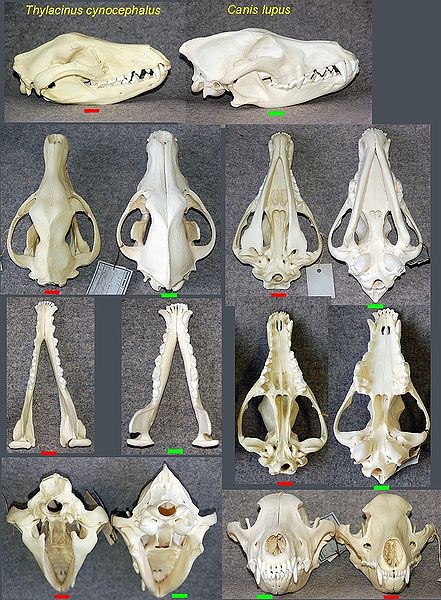
Found this on a creationist website. What do people think?
One of the most fundamental arguments for evolution is the argument from homology, where morphological similarity is used as proof of phyletic relationship due to inheritance from a common ancestor. Classically, symplesiomorphic (shared primitive form) characters were used to group taxa. With the rise of cladistics, however, the focus has shifted to synapomorphic (shared derived form) characters in the determination of phyletic relationships. One of the consequences of that shift is that derived characters that before would be ignored except when dealing with the diagnosis for a particular taxon are now an integral part of the phyletic analysis. This gives a more comprehensive view of the character distributions within a group of taxa.
Homoplasy-correspondence between parts or organs acquired as the result of parallel evolution or convergence. (http://www.merriam-w...onary/homoplasy) Homoplasy is a flat contradiction to the principle of using morphological similarity to prove relationship, because you cannot attribute such similarity to inheritance, but rather it would have had to originate independently. In reality, it is nothing more than an ad hoc explanation as to why the principle of homology doesn't work in a particular instance.
During the 1980's and 1990's several workers including, but not limited to, Richard C. Hulbert Jr., Bruce J. MacFadden, and Thomas S. Kelly, did a comprehensive revision of the mesodont (middle-crowned) and hypsodont (high-crowned) grazing horses. While a college student in the '90s, I mapped out the character distributions in several of their cladistic analyses, and the first of these I am presenting here. In 1989, Hulbert published an analysis that included the 13 well established genera of hypsodont horses, and 10 "Merychippus" grade mesodont horses, with "Parahippus" leonensis as the outgroup (Hulbert, Richard C. Jr., 1989. Phylogenetic Interrelationships and Evolution of North American Late Neogene Equidae. In "The Evolution of Perissodactyls", Prothero, D. R. and Schoch, R. M. (eds.)). He used 45 characters, 7 of which have multiple states (3 with differently derived states, and 4 with sequentially derived states), for a total of 52 derived characters.
Below is Hulbert's fig 11.2 cladogram with all of the characters mapped and color-coded as follows: Yellow= unreversed synapomorphies, i.e. characters that can be unequivocally attributed to shared inheritance. Green= synapomorphies that are subsequently reversed in one or more taxa and are therefore equivocal in there use as proof of relationship. Pink= reversals of the above green characters. Blue= parallelisms where the character would have to have been acquired independently more than once, i.e. homoplasy. Gray= autapomorphic characters unique to a single taxon and therefore useless for determining phyletic relationship.
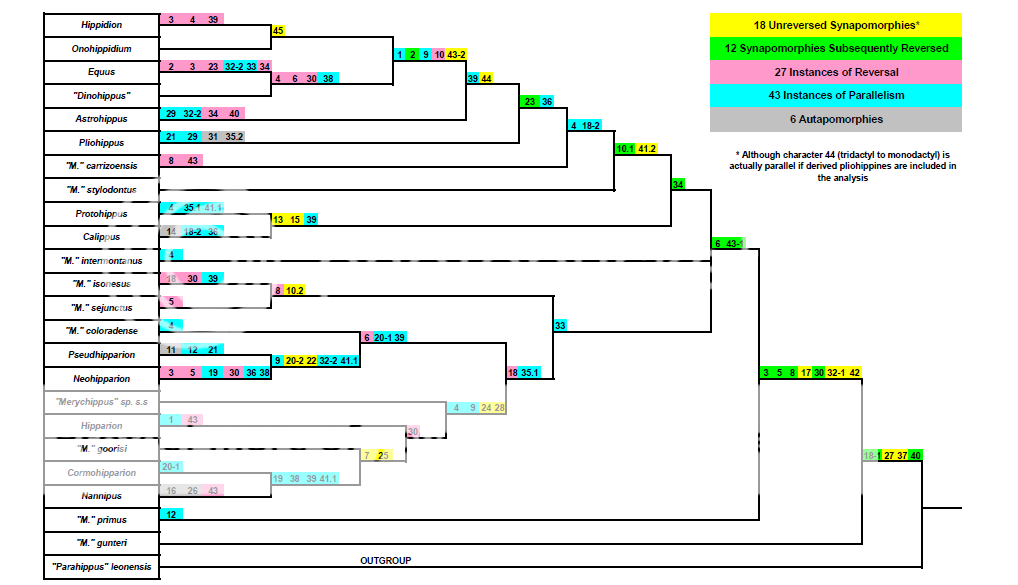
17 of the characters prove to be unreversed synapomorphies and therefore can be used as unequivocal evidence of phyletic descent. I have 18 mapped as yellow, but character 44--tridactyl to monodactyl feet--is actually a parallelism in the Equinae. Hulbert used Pliohippus mirabilis, a tridactyl form, to score the characters for Pliohippus. But the more derived Pliohippus pernix and Pliohippus nobilis are monodactyl (see, for example, Kelly http://www.nhm.org/s...ience/CS473.pdf), so one-toed feet would have had to originate independently twice-once in the Pliohippus clade, and again in the Astrohippus-Dinohippus-Equus-Onohippidium-Hippidion clade.
12 of the characters are synapomorphic but get reversed, and so are equivocal in their use in establishing phyletic relationship.
6 of the characters are autapomorphic (unique to a single taxon) and therefore useless for determining interrelationships.
17 of the characters are parallelisms, or, in other words, homoplastic, and therefore contradict the principle of homology.
A count of the total number of instances where a character state has been changed yields 29 synapomorphies, 27 reversals, and 44 parallelisms (some parallelisms occur more than twice). Now, I'm no mathematical genius, but when the ratio of the number of occurrences that support a method (29 synapomorphies) to the number of occurrences that contradict a method (44 parallelisms) is about 2:3, there is something seriously flawed with the method. If you include the reversals with the parallelisms, it gets even worse, about 2:5.
My question for evolutionists is this: if the practice of using morphological similarity to prove phyletic relationship due to inheritance from a common ancestor performs so poorly and is so equivocal in such a closely (and truly) related group like the Equinae, why should it carry any credibility when dealing with such widely disparate forms like a human, a bat, and a whale? The only way I can see to maintain such an argument is if you engage in extreme "cherry picking" in your selection of characters that "prove" relationship.
It would seem to me that the more reasonable hypothesis, at least with regard to the Equidae, is that all these characters are "subroutines" that were built into the equid genome from the start. And that allowed them to be mosaically expressed in different combinations as the horses spread out and diversified in the rapidly changing early post flood world. Of course, that presents an even bigger problem for an evolutionary origin of these characters. How do you select for and preserve a character that hasn't even been expressed yet?