Rumraket
Active Member
I have to tell this story because irony doesn't get any better than this.
As many of you no doubt know, much of creationist "issues" with evolution revolves around the endlessly debunked analogy of evolution, originally espoused by Fred Hoyle, about a tornado going through a junk-yard and assembling a functional Boeing 747 "by chance". This terrible analogy has been used so much it had it's own fallacy named after it, Hoyle's Fallacy. Over on rationalskepticism.org known by another name, courtesy of Calilasseia, the 'One true sequence-fallacy'.
Anyway, the particular claim of creationists is that, because there are so many possible sequences, it's extremely unlikely to find one that works by chance. Problem was it was always a theoretical argument, so they needed some concrete real-world experiment to really show it. Surely with an experiment, pesky Darwinists couldn't ignore them any more! Back in 2004, ID-creationist scientist, a Chemical Engineer* by the name of Douglas Axe decided he'd be the man to do it.
*(and chemical engineers have no qualifications in evolutionary biology, but whatever...)
He intelligently designed the killer experiment that would demonstrate new functions for protein molecules could not evolve because it was too statistically unlikely. That work is reviewed here: Axe (2004) and the evolution of enzyme function.
To make a long story as short as I can, he took a particular type of enzyme used by bacteria to combat antibiotics, called a Beta-lactamase, and put mutations into it. Beta-lactamase breaks down ampicillin (a beta-lactam), a common antiobiotic used against bacteria.
So he mutated the Beta-lactamase until it stopped working, as in, it stopped breaking down ampicillin. Then he counted how many mutations it took, and with that information he tried to estimate how many functional "versions" of the beta-lactamase enzyme there are, out of the total possible space of all proteins of the same size (150 amino acids long polymer). He came up with a fantastically low number, 1 in 10[sup]77[/sup].
So his claim would be that, out of the total space of proteins that are 150 amino acids long, a space of the size ~1.43x10[sup]195[/sup] (an unfathomably vast number), only 1 in every 10[sup]77[/sup] protein molecules would be functional. While that would still total lots of functional protein molecules in that enormous space, given how many nonfunctional ones there are for every functional one, the odds of ever happening to find a new function with mutations would be so low as to be virtually impossible.
Or in other words, for all intents and purposes, evolution could not be expected to have evolved a single new protein function in the entire history of life (~4 billion years) even if the entire planet was covered in a kilometer thick broth of bacteria.
And he's right, given a few simplifying assumptions (such as if the distribution of function in the space is roughly equidistant, which btw is known to be false). If his number really is correct, with such a low frequency of functional proteins in the total possible space, randomly mutating a functional sequence and happening to find a new one, would be a statistical "miracle".
Now, there are lots of reasons to think his number is wrong (and that his experiment can't be used to reach his conclusion), which I'm not going to go into here. I debunked that before here: Axe, EN&W and protein sequence space (again, again, again).
Instead, I want to tell a story about irony. In 1935 humans invented a synthetic compound never before seen on planet Earth, called nylon. Many of you are familiar with the case of the evolution of a new enzyme called nylonase in the common vernacular, which degrades the waste products of nylon manyfacture. To really appreciate the irony I want to tell you about, we have to look into the history of the discovery of nylonase.
First of all, "nylonase" is not actually a single enzyme. It refers to any one of three enzymes known to react with different nylon manufacture waste-products. In 1975 these enzymes were discovered and called NylA, NylB, and NylC. They all act on different waste products of nylon manufacture. At the time they were known to exist only in a single species of bacteria (today called Flavobacterium), all encoded on the same plasmid (designated 'pOAD2').
The key enzyme required for effective bacterial nylon-waste metabolism is NylB. NylB gradually breaks down nylon polymers (6-aminohexanoate oligomers) into monomers, breaking off one monomer at a time. The other two, NylA and NylC are not required for effective nylon-waste metabolism, they react on other waste-products of nylon, which they in turn convert to substrates that NylB can break down.
The pathway looks like this:
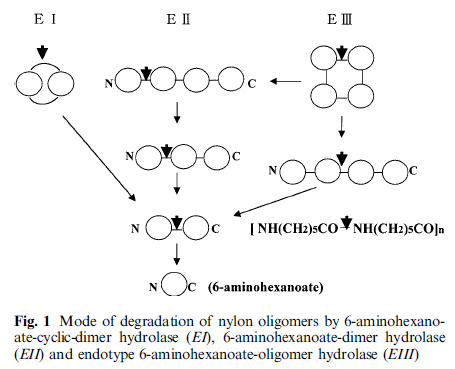
EI = NylA
EII = NylB
EIII = NylC
As you can see, NylB is the important enzyme since it's responsible for the complete breakdown of the nylon oligomer. In 1975 when they were fist discovered, not much was known about NylB (and even less about NylA and NylC, which I'm going to ignore in this post, because short story even shorter, we still don't know how they evolved).
There were no known sequence homologues to any of the three enzymes, so scientists had no idea how they evolved. They also were not similar to each other, so they did not evolve from each other either. All that was known at that time was that a species of bacterium could live in agar containing nylon-waste as the sole carbon source.
Later on, by around 1985 it had become clear that other bacteria could also catalyze the breakdown of nylon oligomers. Interestingly, another species with nylon-waste metabolic ability had a distant homologoue of NylB (only 37% amino acid similarity, no DNA homology) also encoded on one of it's plasmids. A plasmid that was homologous to the pOAD2 plasmid from Flavobacterium. With such a different enzyme (NylB is 392 amino acids long), it was reasoned these two enzymes diverged about 140 million years ago from a common ancestor enzyme with an unknown function.
Here's where it gets interesting. NylB was known from two species of bacteria, Flavobacterium and Pseudomonas (particular strains of these of course, but I'm not going to clutter the post with abbreviations when I can avoid it).
So scientists named the two distantly related enzymes F-NylB and P-NylB (from Flavobacterium and Pseudomonas of course). Additionally, in Flavobacterium there was a slightly different duplicate of the F-NylB gene, which they called F-NylB' (with 88% amino acid sequence similarity to F-NylB).
This gave scientists enough information to be able to give a rough estimate of the particular evolutionary history of nylon metabolism, both before and subsequent to 1935.
The story goes something like this:
140 million years ago, an ancestor of flavobacterium carried a plasmid with a gene encoding a protein of an unknown function, 472 amino acids long. A frameshift mutation created an alternate reading frame in this gene, resulting in a 392 amino acid protein, also of unknown function. This protein turned out to be beneficial to the organism, so it was retained.
The plasmid carrying this protein was horizontally transferred to the Pseudomonas bacterium (or from Pseudomonas to Flavobacterium, which one had the original isn't known and doesn't actually matter). Then the "proto"-NylB gene diverged independently in both organisms. It was still retained in both because it was apparently useful, but it also mutated a lot independently in both lineages, so much that eventually it's less than 40% similar in amino acid sequence today.
Eventually over a hundred million years after the horizontal transfer, the proto-NylB gene in Flavobacterium gets duplicated so there is "proto"-NylB and "proto"-NylB'.
Millions of years more pass, during which time proto-NylB and proto-NylB' diverge by about 12% in amino acid sequence (that's about 47 amino acids out of the 392 total).
Eventually Flavobacterium finds itself in a waste-water pipe of the worlds first nylon factory in 1935. Coincidentally, the F-NylB enzyme, which is 88% similar to F-NylB', can catalyze the breakdown of the nylon waste. Between 1935 and 1975, F-NylB takes on two additional amino acid substitutions, so it is now 200 times better at catalyzing nylon waste breakdown than it was in 1935. F-NylB' is still 200 times worse at catalyzing nylon waste breakdown compared to F-NylB, which implies it is retained to act on the "ancestral" function to nylon-metabolism.
I made a timeline:
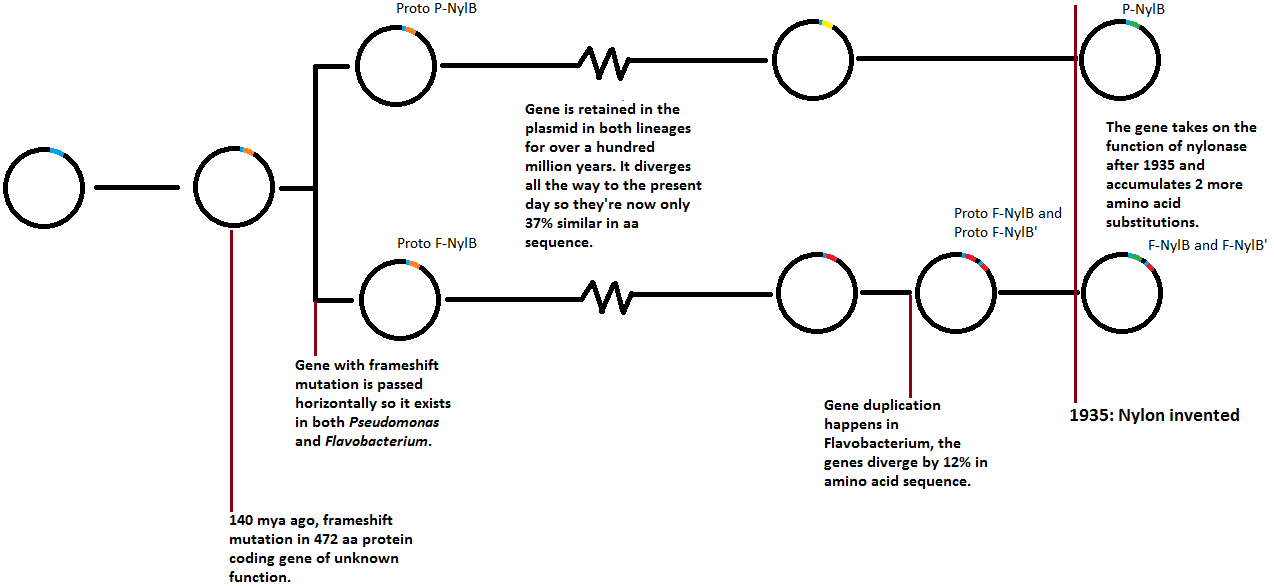
Between 1984 and 1992, scientists work out the above history for the NylB enzymes and it's homologues. They still don't know what the hell NylB did before nylon evolved. All they know is it must have been useful to have been retained all the way from it's origin in a frameshift mutation 140 million years ago, otherwise it should have been lost to the accumulation of deletion mutations.
What was weird about it was that NylB belongs to a particular class of enzymes called amide hydrolases. That is the particular class of chemical reactions it catalyzes. The hydrolysis of amide-bonds.
Naturally, scientists reasoned it must have evolved from other amide-hydrolases. They tested it on literally hundreds of natural amide compounds back in the 1990's, but it worked on none of them. Not even a little. It ONLY worked on the amide bond in nylon as far as they could gather.
Eventually, 2005 comes along and some scientists want to find out what the hell kind of enzyme NylB evolved from. So they work to resolve the structure of the enzyme to see if maybe they can determine from it's structure what it used to do, before nylon was invented by humans.
... they find out it's.. a Beta-lactamase. It catalyzes the break-down of ampicillin (a beta-lactam). Particularly a type of chemical reaction called carboxylic ester hydrolysis. Which means it hydrolyses ester-bonds.
When some plants or fungi once upon a time evolved the beta-lactam antibiotics over 140 million years ago, bacteria in turn evolved resistance to it. There evolved a beta-lactamase enzyme, de novo, by a frameshift mutation in another gene.
Beta-lactamase. The very kind of enzyme that Douglas Axe used, in 2004, to try to prove that evolution of new enzyme functions is statistically impossible.
Irony, your name is beta-lactamase.
Some references:
Shinichi KINOSHITA, Seiji NEGORO, Minoru MURAMATSU, V. S. BISARIA, Shujo SAWADA, Hirosuke OKADA. 6-Aminohexanoic Acid Cyclic Dimer Hydrolase. A New Cyclic Amide Hydrolase Produced by Acromobacter guttatus KI72. Eur J Biochem. 1977 Nov 1;80(2):489-95. DOI: 10.1111/j.1432-1033.1977.tb11904.x
Ohno, S. 1984. Birth of a Unique Enzyme from an Alternative Reading Frame of the Preexisted, Internally Repetitious Coding Sequence. Proceedings, National Academy of Sciences 81:2421-2425. PMCID: PMC345072
Negoro S, Kakudo S, Urabe I, Okada H. A new nylon oligomer degradation gene (nylC) on plasmid pOAD2 from a Flavobacterium sp. J Bacteriol. 1992 Dec;174(24):7948-53. PMCID: PMC207530
Kanagawa K, Oishi M, Negoro S, Urabe I, Okada H. Characterization of the 6-aminohexanoate-dimer hydrolase from Pseudomonas sp. NK87. J Gen Microbiol. 1993 Apr;139(4):787-95. DOI: 10.1099/00221287-139-4-787
Negoro S. Biodegradation of nylon oligomers. Appl Microbiol Biotechnol (2000) 54: 461. doi: 10.1007/s002530000434
Negoro S1, Ohki T, Shibata N, Mizuno N, Wakitani Y, Tsurukame J, Matsumoto K, Kawamoto I, Takeo M, Higuchi Y. X-ray crystallographic analysis of 6-aminohexanoate-dimer hydrolase: molecular basis for the birth of a nylon oligomer-degrading enzyme. J Biol Chem. 2005 Nov 25;280(47):39644-52. Epub 2005 Sep 14. DOI: 10.1074/jbc.M505946200
As many of you no doubt know, much of creationist "issues" with evolution revolves around the endlessly debunked analogy of evolution, originally espoused by Fred Hoyle, about a tornado going through a junk-yard and assembling a functional Boeing 747 "by chance". This terrible analogy has been used so much it had it's own fallacy named after it, Hoyle's Fallacy. Over on rationalskepticism.org known by another name, courtesy of Calilasseia, the 'One true sequence-fallacy'.
Anyway, the particular claim of creationists is that, because there are so many possible sequences, it's extremely unlikely to find one that works by chance. Problem was it was always a theoretical argument, so they needed some concrete real-world experiment to really show it. Surely with an experiment, pesky Darwinists couldn't ignore them any more! Back in 2004, ID-creationist scientist, a Chemical Engineer* by the name of Douglas Axe decided he'd be the man to do it.
*(and chemical engineers have no qualifications in evolutionary biology, but whatever...)
He intelligently designed the killer experiment that would demonstrate new functions for protein molecules could not evolve because it was too statistically unlikely. That work is reviewed here: Axe (2004) and the evolution of enzyme function.
To make a long story as short as I can, he took a particular type of enzyme used by bacteria to combat antibiotics, called a Beta-lactamase, and put mutations into it. Beta-lactamase breaks down ampicillin (a beta-lactam), a common antiobiotic used against bacteria.
So he mutated the Beta-lactamase until it stopped working, as in, it stopped breaking down ampicillin. Then he counted how many mutations it took, and with that information he tried to estimate how many functional "versions" of the beta-lactamase enzyme there are, out of the total possible space of all proteins of the same size (150 amino acids long polymer). He came up with a fantastically low number, 1 in 10[sup]77[/sup].
So his claim would be that, out of the total space of proteins that are 150 amino acids long, a space of the size ~1.43x10[sup]195[/sup] (an unfathomably vast number), only 1 in every 10[sup]77[/sup] protein molecules would be functional. While that would still total lots of functional protein molecules in that enormous space, given how many nonfunctional ones there are for every functional one, the odds of ever happening to find a new function with mutations would be so low as to be virtually impossible.
Or in other words, for all intents and purposes, evolution could not be expected to have evolved a single new protein function in the entire history of life (~4 billion years) even if the entire planet was covered in a kilometer thick broth of bacteria.
And he's right, given a few simplifying assumptions (such as if the distribution of function in the space is roughly equidistant, which btw is known to be false). If his number really is correct, with such a low frequency of functional proteins in the total possible space, randomly mutating a functional sequence and happening to find a new one, would be a statistical "miracle".
Now, there are lots of reasons to think his number is wrong (and that his experiment can't be used to reach his conclusion), which I'm not going to go into here. I debunked that before here: Axe, EN&W and protein sequence space (again, again, again).
Instead, I want to tell a story about irony. In 1935 humans invented a synthetic compound never before seen on planet Earth, called nylon. Many of you are familiar with the case of the evolution of a new enzyme called nylonase in the common vernacular, which degrades the waste products of nylon manyfacture. To really appreciate the irony I want to tell you about, we have to look into the history of the discovery of nylonase.
First of all, "nylonase" is not actually a single enzyme. It refers to any one of three enzymes known to react with different nylon manufacture waste-products. In 1975 these enzymes were discovered and called NylA, NylB, and NylC. They all act on different waste products of nylon manufacture. At the time they were known to exist only in a single species of bacteria (today called Flavobacterium), all encoded on the same plasmid (designated 'pOAD2').
The key enzyme required for effective bacterial nylon-waste metabolism is NylB. NylB gradually breaks down nylon polymers (6-aminohexanoate oligomers) into monomers, breaking off one monomer at a time. The other two, NylA and NylC are not required for effective nylon-waste metabolism, they react on other waste-products of nylon, which they in turn convert to substrates that NylB can break down.
The pathway looks like this:
EI = NylA
EII = NylB
EIII = NylC
As you can see, NylB is the important enzyme since it's responsible for the complete breakdown of the nylon oligomer. In 1975 when they were fist discovered, not much was known about NylB (and even less about NylA and NylC, which I'm going to ignore in this post, because short story even shorter, we still don't know how they evolved).
There were no known sequence homologues to any of the three enzymes, so scientists had no idea how they evolved. They also were not similar to each other, so they did not evolve from each other either. All that was known at that time was that a species of bacterium could live in agar containing nylon-waste as the sole carbon source.
Later on, by around 1985 it had become clear that other bacteria could also catalyze the breakdown of nylon oligomers. Interestingly, another species with nylon-waste metabolic ability had a distant homologoue of NylB (only 37% amino acid similarity, no DNA homology) also encoded on one of it's plasmids. A plasmid that was homologous to the pOAD2 plasmid from Flavobacterium. With such a different enzyme (NylB is 392 amino acids long), it was reasoned these two enzymes diverged about 140 million years ago from a common ancestor enzyme with an unknown function.
Here's where it gets interesting. NylB was known from two species of bacteria, Flavobacterium and Pseudomonas (particular strains of these of course, but I'm not going to clutter the post with abbreviations when I can avoid it).
So scientists named the two distantly related enzymes F-NylB and P-NylB (from Flavobacterium and Pseudomonas of course). Additionally, in Flavobacterium there was a slightly different duplicate of the F-NylB gene, which they called F-NylB' (with 88% amino acid sequence similarity to F-NylB).
This gave scientists enough information to be able to give a rough estimate of the particular evolutionary history of nylon metabolism, both before and subsequent to 1935.
The story goes something like this:
140 million years ago, an ancestor of flavobacterium carried a plasmid with a gene encoding a protein of an unknown function, 472 amino acids long. A frameshift mutation created an alternate reading frame in this gene, resulting in a 392 amino acid protein, also of unknown function. This protein turned out to be beneficial to the organism, so it was retained.
The plasmid carrying this protein was horizontally transferred to the Pseudomonas bacterium (or from Pseudomonas to Flavobacterium, which one had the original isn't known and doesn't actually matter). Then the "proto"-NylB gene diverged independently in both organisms. It was still retained in both because it was apparently useful, but it also mutated a lot independently in both lineages, so much that eventually it's less than 40% similar in amino acid sequence today.
Eventually over a hundred million years after the horizontal transfer, the proto-NylB gene in Flavobacterium gets duplicated so there is "proto"-NylB and "proto"-NylB'.
Millions of years more pass, during which time proto-NylB and proto-NylB' diverge by about 12% in amino acid sequence (that's about 47 amino acids out of the 392 total).
Eventually Flavobacterium finds itself in a waste-water pipe of the worlds first nylon factory in 1935. Coincidentally, the F-NylB enzyme, which is 88% similar to F-NylB', can catalyze the breakdown of the nylon waste. Between 1935 and 1975, F-NylB takes on two additional amino acid substitutions, so it is now 200 times better at catalyzing nylon waste breakdown than it was in 1935. F-NylB' is still 200 times worse at catalyzing nylon waste breakdown compared to F-NylB, which implies it is retained to act on the "ancestral" function to nylon-metabolism.
I made a timeline:
Between 1984 and 1992, scientists work out the above history for the NylB enzymes and it's homologues. They still don't know what the hell NylB did before nylon evolved. All they know is it must have been useful to have been retained all the way from it's origin in a frameshift mutation 140 million years ago, otherwise it should have been lost to the accumulation of deletion mutations.
What was weird about it was that NylB belongs to a particular class of enzymes called amide hydrolases. That is the particular class of chemical reactions it catalyzes. The hydrolysis of amide-bonds.
Naturally, scientists reasoned it must have evolved from other amide-hydrolases. They tested it on literally hundreds of natural amide compounds back in the 1990's, but it worked on none of them. Not even a little. It ONLY worked on the amide bond in nylon as far as they could gather.
Eventually, 2005 comes along and some scientists want to find out what the hell kind of enzyme NylB evolved from. So they work to resolve the structure of the enzyme to see if maybe they can determine from it's structure what it used to do, before nylon was invented by humans.
... they find out it's.. a Beta-lactamase. It catalyzes the break-down of ampicillin (a beta-lactam). Particularly a type of chemical reaction called carboxylic ester hydrolysis. Which means it hydrolyses ester-bonds.
When some plants or fungi once upon a time evolved the beta-lactam antibiotics over 140 million years ago, bacteria in turn evolved resistance to it. There evolved a beta-lactamase enzyme, de novo, by a frameshift mutation in another gene.
Beta-lactamase. The very kind of enzyme that Douglas Axe used, in 2004, to try to prove that evolution of new enzyme functions is statistically impossible.
Irony, your name is beta-lactamase.
Some references:
Shinichi KINOSHITA, Seiji NEGORO, Minoru MURAMATSU, V. S. BISARIA, Shujo SAWADA, Hirosuke OKADA. 6-Aminohexanoic Acid Cyclic Dimer Hydrolase. A New Cyclic Amide Hydrolase Produced by Acromobacter guttatus KI72. Eur J Biochem. 1977 Nov 1;80(2):489-95. DOI: 10.1111/j.1432-1033.1977.tb11904.x
Ohno, S. 1984. Birth of a Unique Enzyme from an Alternative Reading Frame of the Preexisted, Internally Repetitious Coding Sequence. Proceedings, National Academy of Sciences 81:2421-2425. PMCID: PMC345072
Negoro S, Kakudo S, Urabe I, Okada H. A new nylon oligomer degradation gene (nylC) on plasmid pOAD2 from a Flavobacterium sp. J Bacteriol. 1992 Dec;174(24):7948-53. PMCID: PMC207530
Kanagawa K, Oishi M, Negoro S, Urabe I, Okada H. Characterization of the 6-aminohexanoate-dimer hydrolase from Pseudomonas sp. NK87. J Gen Microbiol. 1993 Apr;139(4):787-95. DOI: 10.1099/00221287-139-4-787
Negoro S. Biodegradation of nylon oligomers. Appl Microbiol Biotechnol (2000) 54: 461. doi: 10.1007/s002530000434
Negoro S1, Ohki T, Shibata N, Mizuno N, Wakitani Y, Tsurukame J, Matsumoto K, Kawamoto I, Takeo M, Higuchi Y. X-ray crystallographic analysis of 6-aminohexanoate-dimer hydrolase: molecular basis for the birth of a nylon oligomer-degrading enzyme. J Biol Chem. 2005 Nov 25;280(47):39644-52. Epub 2005 Sep 14. DOI: 10.1074/jbc.M505946200

