AronRa
Administrator
There is a video where a pastor is defending Roy Moore. So I commented, "A pastor saying "You can't say it like it's fact". How ironic. All religion does is assert empty unsupported speculation as if it were fact." My comment got the attention of a creationist commenter.
Just for clarification, a belief-system has required beliefs and prohibited beliefs, but free thought is exactly opposite of that. It doesn't matter what you believe, all that matters is why you believe it.
So the very first thing Owens said was false. His second sentence was false too.
my·thol·o·gy
/məˈTHäləjē/
noun
1. a collection of myths, especially one belonging to a particular religious or cultural tradition.
"Ganesa was the god of wisdom and success in Hindu mythology"
synonyms: myth(s), legend(s), folklore, folk tales, folk stories, lore, tradition
"no ancient culture is without its mythology"
2. the study of myths.
myth
/miTH/
noun
1. a traditional story, especially one concerning the early history of a people or explaining some natural or social phenomenon, and typically involving supernatural beings or events.
synonyms: folk tale, folk story, legend, tale, story, fable, saga, mythos, lore, folklore, mythology
"ancient Greek myths"
2. a widely held but false belief or idea.
"he wants to dispel the myth that sea kayaking is too risky or too strenuous"
Science is neither a tradition nor a story. It's an investigation, which means it changes according to indications of evidence, where his mythology can't. Science doesn't involve the supernatural either because it has to be testable and potentially falsifiable. As I told this guy before, whatever I say has to be verifiably accurate or it's wrong, but whatever believers say is at best unsupported assertions. So they're accidentally false when they're not deliberately false. What my critic has done is put himself on equal standing with me. If I have to produce scientific evidence of my position, then he has to do the same for his own.
Creationists also commonly argue that "we both have the same evidence", which of course isn't true either. Evidence is a body of objectively verifiable facts which are positively indicative of, or exclusively concordant with one available conclusion over any other. If the same fact is still true in either scenario then it isn't evidence. It doesn't become evidence until it aligns with only one of the options over the others. I will show Owens the evidence of my position and he will not be able to contest any of it, nor will he be able to produce any evidence in favor of his own position.
So we don't have the same evidence, but we do have the same observation to explain. In this case, that observation is biodiversity. Why is the tree of life structured as it is? Why do we have all these different breeds of dogs and pigeons, cats and cattle and so on? Why are there taxonomic hierarchies beyond that between different parent groupings of lizards, as there are for birds and everything else? If Owens were sincere in his position, he would take the Phylogeny Challenge. But as he is a creationist, he will not take that challenge because he is not sincere. He only wants to make-believe.
So I'll have to explain to Owens how evolution offers the only explanation for all these and many other things, while he will be unable to show how his mythology can explain any of it.
We also know that humans are a subset of apes, primates, eutherian mammals, and vertebrate deuterostome animals.This fact was observed by many naturalists and philosophers long before Darwin, but most notably by Carolus Linneaus who devised taxonomy, the systematic classification of life.
"Why I demand of you, and of the whole world, that you show me a generic character -one that is according to the generally accepted principles of classification- by which to distinguish between Man and Ape. I myself most assuredly know of none. I wish somebody would indicate one to me. But, if I had called man an ape, or vice versa, I would have fallen under the ban of all ecclesiastics. It may be that as a naturalist, I ought to have done so."
282 years later and still no one can answer that question, because in fact humans ARE apes: meaning that we are part of the superfamily, Hominoidea.
Ardipithecus ramidus kadabba (5.8-5.2 mya) (5 sites, 5 individuals, 0 crania)
Ardipithecus ramidus ramidus (4.4 mya) (2 sites, >50 individuals, 0 crania)
Australopithecus afarensis (4.2-2.96 mya) (11 sites, ~120 individuals,>8 crania)
Australopithecus africanus (2.9-2.4 mya) (7 sites, ~130 individuals, 25 crania)
Australopithecus anamensis (4.17-3.9 mya) (2 sites, 10 individuals, 0 crania)
Australopithecus bahrelghazali (3.5-3.0 mya) (1 site, 1 individual, 0 crania)
Australopithecus garhi (2.5 mya, 3 sites, 4 individuals, 1 cranium)
Homo antecessor (800 kya, 1 site, >5 individuals, 0? crania)
Homo erectus/ergaster (1.9-0.4 mya, >34 sites, >210 individuals, >62 crania)
Homo habilis (2.3-1.6 mya, 7 sites, 25 individuals, 11 crania)
Homo heidelbergensis (700-100 kya, 26 sites, 60 individuals, 20 crania)
Homo sapiens neanderthalensis (250-25 kya, >31 sites, >77 individuals,>27 crania
Homo sapiens sapiens, >10 kya only (130 kya to recent, >75 sites, >154 individuals, numerous crania)
Homo rudolfensis (1.9 mya, 2 sites, 5 individuals, 3 crania)
Kenyanthropus platyops (3.3 mya, 1 site, 3 individuals, 1 crania)
Orrorin tugenensis (6.3-5.6 mya) (4 sites, 5 individuals, 0 crania)
Paranthropus aethiopicus (2.7-1.9 mya, 2 sites, 8 individuals, 2 crania)
Paranthropus boisei (2.5-1.4 mya, 9 sites, 43 individuals, 13 crania)
Paranthropus robustus (2.0-1.5 mya, 3 sites, 28 individuals, 20 crania)
Sahelanthropus tchadensis (7.0-6.0 mya) (1 site, 6 individuals, 1 cranium)
based on this limited data, we have nearly 1,000 individuals, with nearly 200 crania. I expect the final number to be over 3,000 given the estimate of the Catalogue ofFossilHominid published by the British Natural History Museum (almost 4000 as of 1976). It is unclear if this refers to NISP or MNI.
The consensus is that all of these are both extinct apes and extinct hominines. Most of these were predicted by evolutionary theory and should ONLY exist if evolution is true. None of these would exist if creationism were true. How would you--as a creationist--explain extinct humans?
Of course creationists predicted that none of these things would be discovered. Then when they were, creationists had to start lying, saying that Lucy was "just a chimpanzee" or that Neanderthal man was "just an old modern man with rickets". It doesn't matter that every paleoanthropologist knows that Australopithecines were not chimpanzees, nor that we've found hundreds of individual neanderthals including women and children too. When you have a belief system, you have to deny the facts to keep convincing yourself of what you already know can't really be true. That's what religion does, and that's why science is the very opposite.
Dryopithecus is a very broad genus including at least forty species as diverse as gibbon-sized to gorilla-sized, but not actually gibbons or gorillas. Dryopithecines are hominids [family Hominidae] that came before Homininae. The oldest fossils are in Eurasia, and the genus, Pongo [orangutans et al] is Asian, indicating that although Dryopithecus fontana was found in eastern Africa, where the genus Homininae evidently evolved, their ancestors and the crown of Hominidae actually came from Asia through Eurasia.
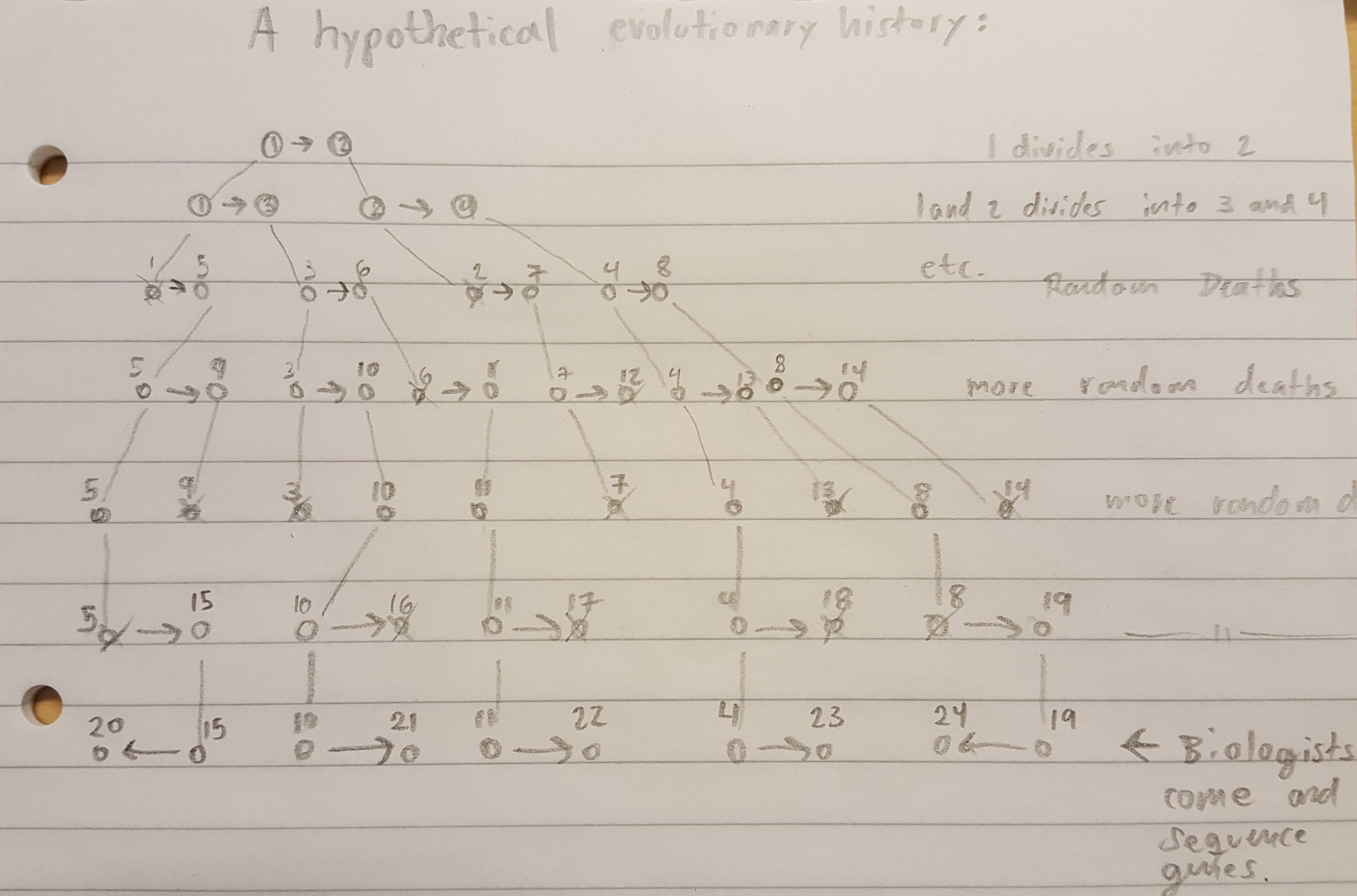
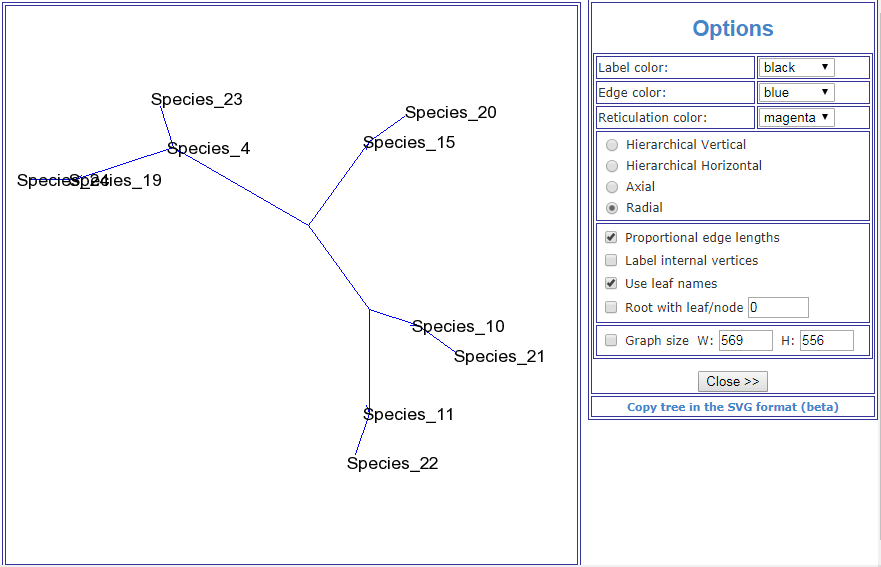
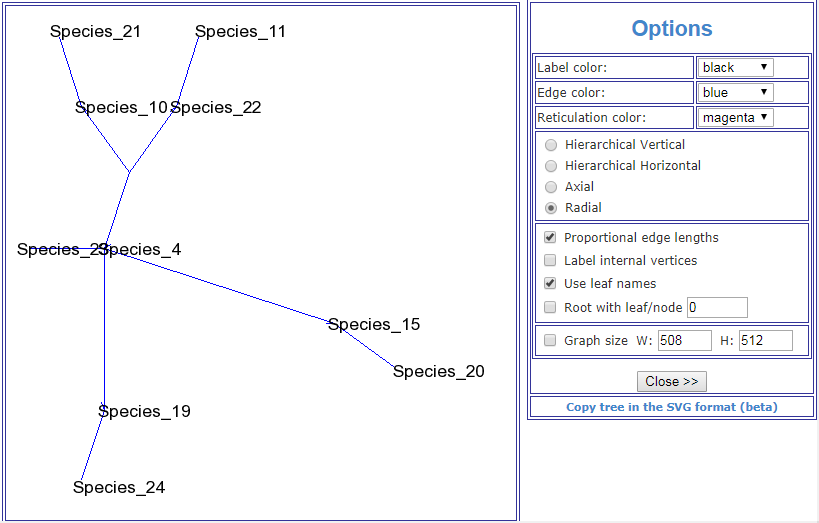
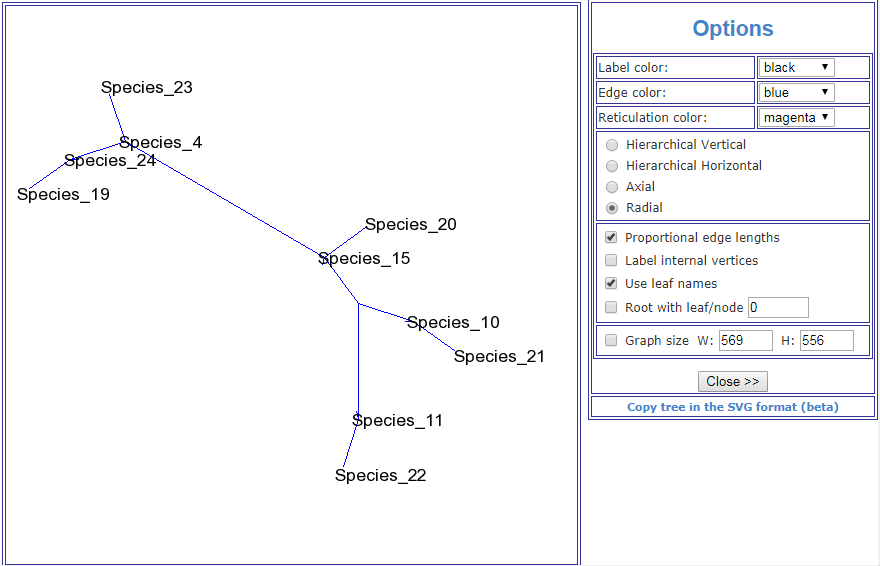
This image is taken from the Public Library of Science article, A Molecular Phylogeny of Living Primates.
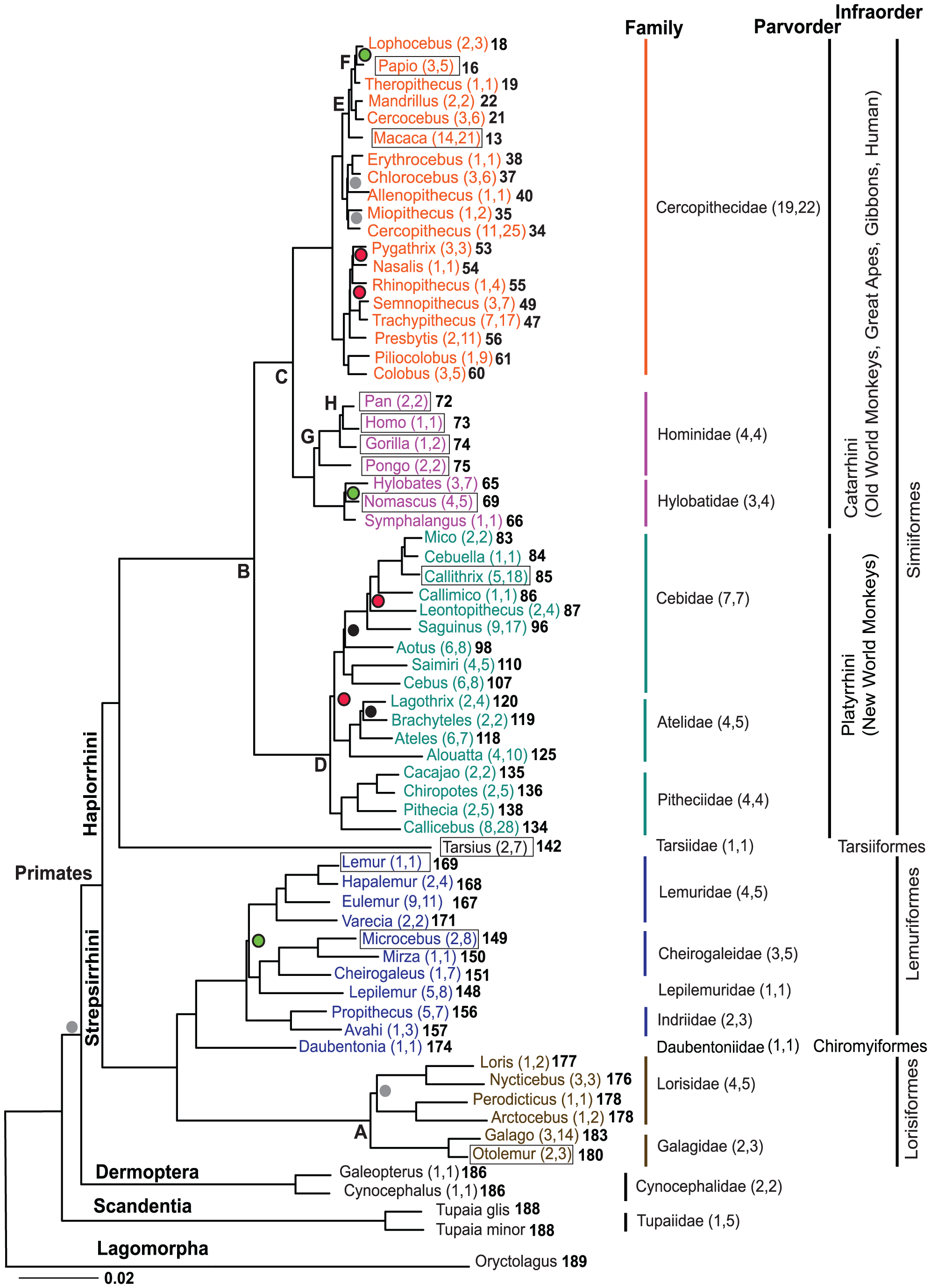
"Here we provide new genomic sequence (∼8 Mb) from 186 primates representing 61 (∼90%) of the described genera, and we include outgroup species from Dermoptera, Scandentia, and Lagomorpha. The resultant phylogeny is exceptionally robust and illuminates events in primate evolution from ancient to recent, clarifying numerous taxonomic controversies and providing new data on human evolution. Ongoing speciation, reticulate evolution, ancient relic lineages, unequal rates of evolution, and disparate distributions of insertions/deletions among the reconstructed primate lineages are uncovered. Our resolution of the primate phylogeny provides an essential evolutionary framework with far-reaching applications including: human selection and adaptation, global emergence of zoonotic diseases, mammalian comparative genomics, primate taxonomy, and conservation of endangered species."
Scientific evidence can't really get any more conclusive than that. So much for your "ZERO evidence" accusation.
You understand that this is a fact, (or a rather a series of them) that was predicted by evolutionary theory because it only aligns with evolution and clearly contradicts creationism, right? Because creationists predicted this would not turn out to be the case. They were wrong again as always. Every prediction falsified.
If evolution is true, what observation, discovery, or experiment could reveal that?
If evolution is not true, then what observation, discovery, or experiment do you predict would reveal that?
If creationism is not true, what observation, discovery, or experiment do you predict could show that?
If creationism is true, what observation, discovery, or experiment could reveal that?
Creationists often use the logical fallacy of false equivalence, asserting that faith is based on evidence just like science is and science relies on faith just like religion does. So "you're just as bad as me and I'm just as good as you." Owen's version of this is to assert that my science is merely mythology just like his religion is. This is also the tu quoquee fallacy, because he asserts that science is a belief-system. Believers must know how dishonest their position is because they keep trying to project their faults onto those who will not share them.Owens said:AronRa, anyone who is honest and has heard you talk about your belief system. knows you do the exact same thing.As I recall, you believe in a mythological common ancestor of man and chimp.Aron Ra said:No, the things I say are verifiably accurate.
Just for clarification, a belief-system has required beliefs and prohibited beliefs, but free thought is exactly opposite of that. It doesn't matter what you believe, all that matters is why you believe it.
So the very first thing Owens said was false. His second sentence was false too.
my·thol·o·gy
/məˈTHäləjē/
noun
1. a collection of myths, especially one belonging to a particular religious or cultural tradition.
"Ganesa was the god of wisdom and success in Hindu mythology"
synonyms: myth(s), legend(s), folklore, folk tales, folk stories, lore, tradition
"no ancient culture is without its mythology"
2. the study of myths.
myth
/miTH/
noun
1. a traditional story, especially one concerning the early history of a people or explaining some natural or social phenomenon, and typically involving supernatural beings or events.
synonyms: folk tale, folk story, legend, tale, story, fable, saga, mythos, lore, folklore, mythology
"ancient Greek myths"
2. a widely held but false belief or idea.
"he wants to dispel the myth that sea kayaking is too risky or too strenuous"
Science is neither a tradition nor a story. It's an investigation, which means it changes according to indications of evidence, where his mythology can't. Science doesn't involve the supernatural either because it has to be testable and potentially falsifiable. As I told this guy before, whatever I say has to be verifiably accurate or it's wrong, but whatever believers say is at best unsupported assertions. So they're accidentally false when they're not deliberately false. What my critic has done is put himself on equal standing with me. If I have to produce scientific evidence of my position, then he has to do the same for his own.
Creationists also commonly argue that "we both have the same evidence", which of course isn't true either. Evidence is a body of objectively verifiable facts which are positively indicative of, or exclusively concordant with one available conclusion over any other. If the same fact is still true in either scenario then it isn't evidence. It doesn't become evidence until it aligns with only one of the options over the others. I will show Owens the evidence of my position and he will not be able to contest any of it, nor will he be able to produce any evidence in favor of his own position.
So we don't have the same evidence, but we do have the same observation to explain. In this case, that observation is biodiversity. Why is the tree of life structured as it is? Why do we have all these different breeds of dogs and pigeons, cats and cattle and so on? Why are there taxonomic hierarchies beyond that between different parent groupings of lizards, as there are for birds and everything else? If Owens were sincere in his position, he would take the Phylogeny Challenge. But as he is a creationist, he will not take that challenge because he is not sincere. He only wants to make-believe.
So I'll have to explain to Owens how evolution offers the only explanation for all these and many other things, while he will be unable to show how his mythology can explain any of it.
He is not talking about the morphological "missing link" that was predicted by Darwin and discovered in 1974 (and many more times since then). He's talking about the common ancestor of that and of chimpanzees. He's talking about Dryopithecus fontana.You have no scientific name for this mythological creature.
Owens actually provided a scientific description for me with his immediate citation of an article in Scientific American, "Fossil Reveals What Last Common Ancestor of Humans and Apes Looked Liked". It was so nice of him to provide the evidence proving that I'm right and he's wrong before we even begin.You cannot even give a scientific description of this mythological creature.
Because all the evidence, including the bit you just cited supports it. That article confessed that they didn't have enough of the skull to build a facial restoration. So we can't yet say what it looked like, (other than it looked vaguely like a gibbon) but that doesn't mean it didn't exist, or that the fragments of it don't match subsequent Hominids.How is that not unsupported speculation?
Don't we? We know that evolution happens, that significant beneficial mutations do occur and are inherited by descendant groups, and that multiple independent sets of biological markers exist to trace these lineages backward over many generations. We know that the collective genome of all animals has been traced to its most basal form, and that those forms are also indicated by comparative morphology, physiology, and embryological development. Consequently we know that everything on earth has definite relatives either living nearby or evident in the fossil record, and we know that the fossil record holds hundreds of transitional species even according to its strictest definition. We also know that both microevolution and macroevolution have been directly observed, and that evolutionary variation is pervasive and ubiquitous throughout the biology. That's why evolution is the basis of modern biology.what is your evidence that Dryopithecus fontana is the common ancetor of both humans and apes?We don't have evidence confirming that we are evolved apes.So once we had the evidence confirming that we are evolved apes, we then needed evidence of intermediate connections between us and them. We now have a couple dozen species representing different stages of that transition, and none of them would exist under the creationist model.
Then once we knew our origin among the apes, we needed evidence of the origin of the other modern apes too. Once we established the Dryopithecines are basal to both modern apes and humans, the problem was that they're European. So we needed evidence of Dryopithecines in east Africa, and now we have it. Dryopithecus fontana.
What more evidence could we possibly need that we don't already have?
We also know that humans are a subset of apes, primates, eutherian mammals, and vertebrate deuterostome animals.This fact was observed by many naturalists and philosophers long before Darwin, but most notably by Carolus Linneaus who devised taxonomy, the systematic classification of life.
"Why I demand of you, and of the whole world, that you show me a generic character -one that is according to the generally accepted principles of classification- by which to distinguish between Man and Ape. I myself most assuredly know of none. I wish somebody would indicate one to me. But, if I had called man an ape, or vice versa, I would have fallen under the ban of all ecclesiastics. It may be that as a naturalist, I ought to have done so."
282 years later and still no one can answer that question, because in fact humans ARE apes: meaning that we are part of the superfamily, Hominoidea.
Wrong. Here is a really old list that needs updating.Nor do we have evidence of intermediate connections. What we do have are either extinct apes or extinct humans based on a few questionable bone fragments. There is no consensus on any of the so-called intermediate stages.
Ardipithecus ramidus kadabba (5.8-5.2 mya) (5 sites, 5 individuals, 0 crania)
Ardipithecus ramidus ramidus (4.4 mya) (2 sites, >50 individuals, 0 crania)
Australopithecus afarensis (4.2-2.96 mya) (11 sites, ~120 individuals,>8 crania)
Australopithecus africanus (2.9-2.4 mya) (7 sites, ~130 individuals, 25 crania)
Australopithecus anamensis (4.17-3.9 mya) (2 sites, 10 individuals, 0 crania)
Australopithecus bahrelghazali (3.5-3.0 mya) (1 site, 1 individual, 0 crania)
Australopithecus garhi (2.5 mya, 3 sites, 4 individuals, 1 cranium)
Homo antecessor (800 kya, 1 site, >5 individuals, 0? crania)
Homo erectus/ergaster (1.9-0.4 mya, >34 sites, >210 individuals, >62 crania)
Homo habilis (2.3-1.6 mya, 7 sites, 25 individuals, 11 crania)
Homo heidelbergensis (700-100 kya, 26 sites, 60 individuals, 20 crania)
Homo sapiens neanderthalensis (250-25 kya, >31 sites, >77 individuals,>27 crania
Homo sapiens sapiens, >10 kya only (130 kya to recent, >75 sites, >154 individuals, numerous crania)
Homo rudolfensis (1.9 mya, 2 sites, 5 individuals, 3 crania)
Kenyanthropus platyops (3.3 mya, 1 site, 3 individuals, 1 crania)
Orrorin tugenensis (6.3-5.6 mya) (4 sites, 5 individuals, 0 crania)
Paranthropus aethiopicus (2.7-1.9 mya, 2 sites, 8 individuals, 2 crania)
Paranthropus boisei (2.5-1.4 mya, 9 sites, 43 individuals, 13 crania)
Paranthropus robustus (2.0-1.5 mya, 3 sites, 28 individuals, 20 crania)
Sahelanthropus tchadensis (7.0-6.0 mya) (1 site, 6 individuals, 1 cranium)
based on this limited data, we have nearly 1,000 individuals, with nearly 200 crania. I expect the final number to be over 3,000 given the estimate of the Catalogue ofFossilHominid published by the British Natural History Museum (almost 4000 as of 1976). It is unclear if this refers to NISP or MNI.
The consensus is that all of these are both extinct apes and extinct hominines. Most of these were predicted by evolutionary theory and should ONLY exist if evolution is true. None of these would exist if creationism were true. How would you--as a creationist--explain extinct humans?
You know, when you haven't studied a particular subject any at all and consequently have absolutely no idea what you're talking about, you probably shouldn't pretend that you know more than all the world's best educated expert specialists in that field. Because we have extensive evidence for the evolution of apes with way more of those species represented in the fossil record than still exist today.No evidence exists for the evolution of modern apes.
You misspelled--and misrepresented--scientific fact. Science is entirely different than a belief system or mythology in that it is an attempt to improve understanding. The only way to do that is to seek out the flaws in your current perspective and correct them. Science does this with predictive hypotheses. If our current understanding of evolution is true then we should find fossils older than modern humans with traits intermediate between us and other apes. That hypothesis was vindicated again and again and again with the above list of dozens of fossil species.Anyone can line up fossils fragments, and make the specious claim that one evolved into the other. Pure science fiction.
Of course creationists predicted that none of these things would be discovered. Then when they were, creationists had to start lying, saying that Lucy was "just a chimpanzee" or that Neanderthal man was "just an old modern man with rickets". It doesn't matter that every paleoanthropologist knows that Australopithecines were not chimpanzees, nor that we've found hundreds of individual neanderthals including women and children too. When you have a belief system, you have to deny the facts to keep convincing yourself of what you already know can't really be true. That's what religion does, and that's why science is the very opposite.
Do you have an example of that? I bet not because that's never happened, but there's no requirement that when a new species evolves that every member of the parent species had to have died out.Evolutionists do this all the time. They claim something evolved into something else and then get busted when it turns out that one of their so-called intermediates is still alive and well !
The article you cited explains otherwise. How could you have quoted one paragraph ouf of that without reading the rest of it?We have not established that Dryopithecines are basal to booth moderan apes and humans.
Dryopithecus is a very broad genus including at least forty species as diverse as gibbon-sized to gorilla-sized, but not actually gibbons or gorillas. Dryopithecines are hominids [family Hominidae] that came before Homininae. The oldest fossils are in Eurasia, and the genus, Pongo [orangutans et al] is Asian, indicating that although Dryopithecus fontana was found in eastern Africa, where the genus Homininae evidently evolved, their ancestors and the crown of Hominidae actually came from Asia through Eurasia.
Taxonomy was replaced with cladistic phylogenetics. It's a twin-nested hierarchy where the trees constructed on morphology can be confirmed genetically. It's rather like taking a paternity test to confirm who your daddy is. Same principle.You have ZERO evidence to support that claim.
This image is taken from the Public Library of Science article, A Molecular Phylogeny of Living Primates.
"Here we provide new genomic sequence (∼8 Mb) from 186 primates representing 61 (∼90%) of the described genera, and we include outgroup species from Dermoptera, Scandentia, and Lagomorpha. The resultant phylogeny is exceptionally robust and illuminates events in primate evolution from ancient to recent, clarifying numerous taxonomic controversies and providing new data on human evolution. Ongoing speciation, reticulate evolution, ancient relic lineages, unequal rates of evolution, and disparate distributions of insertions/deletions among the reconstructed primate lineages are uncovered. Our resolution of the primate phylogeny provides an essential evolutionary framework with far-reaching applications including: human selection and adaptation, global emergence of zoonotic diseases, mammalian comparative genomics, primate taxonomy, and conservation of endangered species."
Scientific evidence can't really get any more conclusive than that. So much for your "ZERO evidence" accusation.
You understand that this is a fact, (or a rather a series of them) that was predicted by evolutionary theory because it only aligns with evolution and clearly contradicts creationism, right? Because creationists predicted this would not turn out to be the case. They were wrong again as always. Every prediction falsified.
If evolution is true, what observation, discovery, or experiment could reveal that?
If evolution is not true, then what observation, discovery, or experiment do you predict would reveal that?
If creationism is not true, what observation, discovery, or experiment do you predict could show that?
If creationism is true, what observation, discovery, or experiment could reveal that?
So here we are. You were wrong on every assumption/assertion/allegation. Every single sentence you said was wrong on every point therein. Yet you think you once refuted me and that you can do it again? You obviously didn't disprove me then just like you can't now either.That's your belief. But until you provide evidence, it's just an empty assertion.